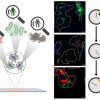
![Targeted proton transfer charge reduction (tPTCR) nano-DESI mass spectrometry imaging of liver tissue from orally dosed rat (Animal 3). a) optical image of a blood vessel within liver tissue. b) Composite ion image of charge-reduced haeme-bound α-globin (7+ and 6+ charge states; m/z 2259.9 and m/z 2636.3 respectively, red) and the charged-reduced [FABP+bezafibrate] complex (7+ and 6+ charge states; m/z 2097.5 and m/z 2446.9 respectively, blue). c) Ion image composed from charge-reduced haeme-bound α-globin (7+ and 6+ charge states) showing abundance in blood vessels. d) Ion image composed from charge-reduced [FABP+bezafibrate] complex (7+ and 6+ charge states) showing abundance in bulk tissue and absence in the blood vessel. Reproduced from https://doi.org/10.1002/ange.202202075 under a CC BY licence. Light and mass spectromert imaging of tissue samples](/sites/default/files/styles/product_photo/public/news/MSI%20drug-protein%20complex-w.jpg?itok=JYHqFmyS)
Researchers at the University of Birmingham, in collaboration with AstraZeneca, have developed a technique that uses native ambient mass spectrometry (MS) to detect intact protein–drug complexes directly from liver tissue.
Part of the early stages of drug discovery takes place in cell cultures, enabling the effects of different compounds to be tested on specific biological targets involved in various diseases. Although this allows researchers to assess how the compounds act against the target, it does not capture the full effects of the physiological environment.
This new technique enables researchers to use real tissue samples to assess which proteins the drug will bind to in the body and, therefore, how effective it is likely to be against the target. Being able to pinpoint the interaction between the drug and the protein can provide valuable insight to guide drug discovery.
“Identifying the drug‒protein interaction at an early stage is incredibly hard. Using MS on proteins is often compared to making an elephant fly. What we’ve done is add an unsecured hat—the drug molecule—to the elephant, and measured the whole process”, said Professor Helen Cooper, School of Biosciences at the University of Birmingham, UK. “Usually in early-stage drug discovery, measurements are taken outside of the physiological environment, so when researchers move onto testing drugs in tissue, they can fail because they have interactions that were not expected.”
In the study, the researchers used tissue taken from the livers of rats dosed with bezafibrate, a drug commonly used to treat high cholesterol. They used MS on thin sections of tissue to detect the drug molecule and the specific fatty acid binding protein to which it attaches to form a complex.
Further, they undertook MS imaging of a dosed liver section by use of native nanospray desorption electrospray ionisation (nano-DESI). This enabled visualisation of the distribution of the [FABP1+bezafibrate] complex throughout the bulk liver tissue and the haeme-bound α-globin in the blood vessels (see image).
Next steps for the research will include improving the sensitivity of the technique, and extending it to other types of drug compounds. Looking further ahead, the team hopes it can be developed for use in human tissue, taken from biopsies. This would yield a greater understanding of why drugs work differently in different patients.