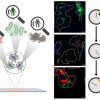
Raman scattering is an inelastic scattering process that exchanges energies between photons and molecules to carry information on molecular vibrations. Raman micro-spectroscopy has become an indispensable analytical tool in biology and medical surgery mainly due to its two “frees”: label-free and water background-free. These benefits enable us to study living samples without endogenous perturbation. In addition, Raman peaks have a much narrower spectral bandwidth than fluorescent dyes’ emission spectrum, which enables the simultaneous study of various metabolic species in the same environment.
In a new paper, Dr Haonan Lin and Professor Ji-Xin Cheng from Boston University reviewed the combination of instrumentation and computational approaches to coherent Raman scattering (CRS). Despite its significant advantages, one fundamental drawback of Raman scattering lies in its severely limited cross-sections. A typical Raman cross-section is 10‒30 cm2 per molecule, resulting in a very long signal integration time from seconds to minutes per focal spot. Such limited speed makes it impractical to perform pixel-by-pixel imaging of dynamic systems. A non-linear optical process has been introduced to enhance the Raman signals coherently and to break the fundamental cross-section limits. With two synchronised ultrafast lasers, coherent Raman signals arose in coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS). In CRS, two laser fields synchronously interact with the target molecule. When the beating frequency matches the Raman vibrational mode, a coherently amplified energy transfer process occurs. It annihilates the pump photon, converts it to the Stokes beam and generates photons at a new frequency.
CRS has enabled high-speed chemical imaging based on intrinsic Raman peaks on biological samples. However, biological samples are sophisticated microsystems that consist of various metabolites which often have spectral overlaps, especially in the strong yet crowded carbon‒hydrogen (CH) region. It hinders the quantitation and identification of chemicals in cells and tissues using narrowband single-colour CRS. Over the past years, significant endeavours have been made to develop hyperspectral CRS that produces a Raman spectrum at each pixel.
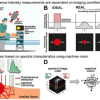
Hyperspectral imaging offers the potential for deciphering the information on chemical compositions and abundance in a complex environment. However, due to the high dimensionality of the raw image, such information is not readily available. Algorithms are required to identify major pure components and decompose concentration maps. Parallel with instrumentation developments in hyperspectral CRS, various hyperspectral image unmixing methods have been reported. Depending on whether prior information is given on the composition of pure components, we categorise them into either supervised or unsupervised methods.
Instrumentation innovations have pushed CRS imaging to the speed of up to 2 kHz frame rate, spectral coverage of up to 3500 cm‒1, and spectral acquisition speed of up to 5 µs per spectrum. However, these conditions cannot be realised simultaneously due to the physical limit determined by the sensitivity limit of CRS. For example, further increasing the speed will deteriorate the setup’s signal-to-noise ratio (SNR), rendering it inapplicable to biomedical applications. Under the constraint of photodamage, this trade-off can be conveyed as a design space. It is a hyperplane intersecting with three axes representing speed, spectral bandwidth and SNR. Optimisation of instrumentation enables the system to reach an optimal condition point on the hyperplane, yet going beyond it remains challenging.
The research team introduced various computational methods used to push the boundary of CRS chemical microscopy. Attention must be paid to the applicable range of computational algorithms to avoid erroneous interpretations of the measurements. Evaluating whether the forward model can appropriately describe the underlying physical process is crucial. It involves the statistical distribution of measurement noise, the image convolution kernel of the imaging system and other techniques.
Rigorous experiments should be taken to characterise the forward model and calibrate model parameters. When prior models/regularisations are used, a comprehensive understanding of the signal is necessary. Hyperparameter tuning for the prior models is crucial for yielding correct results and may require iterative updates and validations. For deep learning applications, although the task of sophisticated modelling on the inverse problem is alleviated, an appropriate selection of network structures and sufficiently large training and validation datasets are necessary.
Looking into the future, the research team expect instrumentation advances will continue to increase the data throughput on temporal, spatial and spectral dimensions. They should provide more features on data structures, such as sparsity and correlation. Meanwhile, new computational methods can be harnessed to break the design space trade-offs and provide enriched chemical compositions for biomedical research. With rapid advances in computational optical microscopy, we expect more ideas to infiltrate CRS.
Since most computational methods focus on wide-field implementations, the translation into CRS microscopy is nontrivial. Extensive modelling, system design and algorithm development need to be performed to ensure applicability to CRS imaging. In the future, computational methods will play an even more critical role as existing methods remain viable to boost the newly established design space. New methods may arise to achieve breakthroughs in aspects such as field of view, imaging depth and spatial resolution.