Antony N. Davies,a,b David Martinsen,c
David Martinsen,c Henry S. Rzepa,d
Henry S. Rzepa,d Charles Romain,d Agustin Barba,e Felipe Seoane,e Santiago Domingueze and Carlos Cobase
Charles Romain,d Agustin Barba,e Felipe Seoane,e Santiago Domingueze and Carlos Cobase
aStrategic Research Group – Measurement and Analytical Science, Akzo Nobel Chemicals b.V., Deventer, the Netherlands
bSERC, Sustainable Environment Research Centre, Faculty of Computing, Engineering and Science, University of South Wales, UK
cDavid Martinsen Consulting, Rockville, MD 20850, USA
dImperial College London, South Kensington Campus, London SW7 2AZ, UK
eMestrelab Research, S.L., Feliciano Barrera 9B – Bajo, 15706 Santiago de Compostela, Spain
DOI: https://doi.org/10.1255/sew.2017.a1
One of the interesting initiatives discussed during the IUPAC General Assembly1 a few weeks ago in Sao Paulo was the renewed push for more efficient and simpler ways of submitting supplementary spectroscopic data. IUPAC Division 3 were particularly keen on enabling better NMR supplementary data submission. It emerged that there have been some interesting efforts made to radically simplify such submission. Mestrelab Research have developed a solution called Mpublish which has been tested and deployed at Imperial College London, aimed at lowering the barriers to the submission of supplementary full spectroscopic data. Along the lines of the EuroSpec project,2,3 the submission system also handles the most complicated data needing submission—multi-dimensional nuclear magnetic resonance spectra. Henry Rzepa’s blog discusses the background and submission process and provides a link to his guide to setting up and uploading files to the Imperial College HPC Data Repository as well as to the “FAIR” principles… (findable, accessible, inter-operable and re-usable).4
Issues to be overcome
As has been discussed extensively, it is crazy that in an internet age, scientists asked to review publications are expected to make decisions on the appropriateness of the work being presented by studying low-resolution images of data. In their day-to-day work in their own labs, the reviewers would have far better tools available to support their decision making. Regulatory compliance agencies are concentrating more on data integrity and the overall desire for better fraud prevention, making available the actual spectroscopic data which is the evidence for claims made within publications is clearly desirable. The benefit for funding bodies has been reflected in, for example, the EU Guidance on access to research data which explains the rules around open access that all beneficiaries of Horizon 2020 funding have to follow.5
However, such a process brings with it a not insignificant amount of additional work for the submitter of a publication. This includes collating, annotating and submitting what—for example in a synthetic organics chemistry paper—could well be quite a large number of files. Not to mention the issue of which format should the electronic data be submitted in—and how to match that submitted with what each reviewer can actually read.
Outline solution
So any new attempt to reduce the barriers to submission of more real supplementary spectroscopic data is to be welcomed. The solution outlined here is the result of a collaborative approach using tools from Mestrelab Research S.L. in a project to see if an end-to-end simplification was possible.
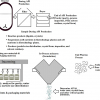
Figure 1 outlines the individual stages that such a solution needs to embody, with each step being more or less independent of one another. Now, this figure is mainly focussed on the submission process, but the results of the peer review can be many and varied. One example cited by Angie Hunter of Organic Letters is that the reviewers may well request the authors to provide more or corrected supplementary information having reviewed the originally submitted paper. The outcome may still not be approval to publish, as the re-submitted paper with or without additional supplementary data can, of course, also fall short of the standard required for a particular journal.
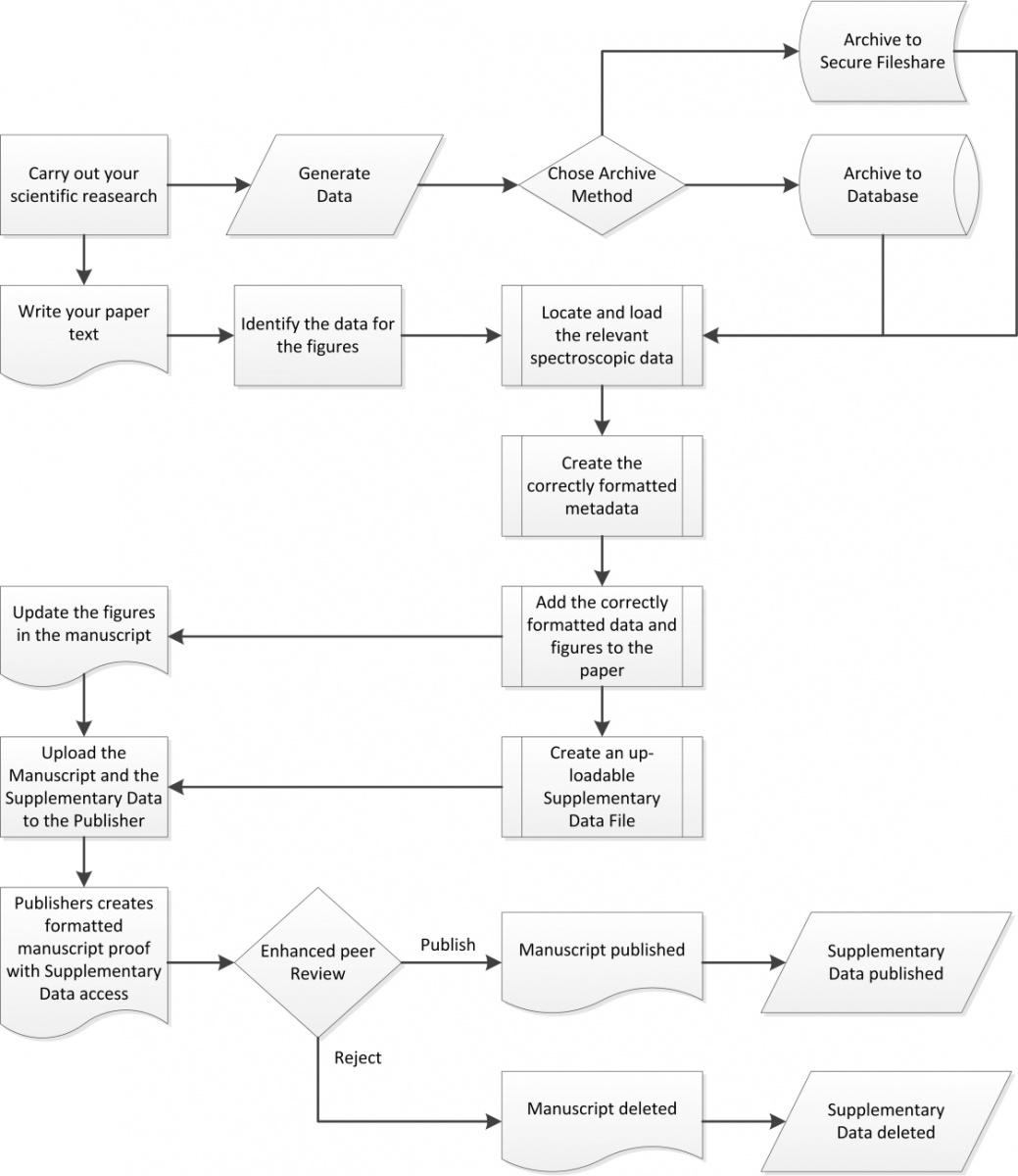
Figure 1. Enhanced publishing workflow with supplementary data capture and submission.
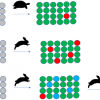
In this solution. they addressed one of the more complex data types. Figure 2 shows a typical example of the spectroscopic information content required for an organic chemistry journal, Organic Letters. Their guidance for authors describes what is required concerning spectra (see “Spectra in manuscript” text box).
Spectra will be published in the body of the manuscript only when concise numerical summaries are inadequate for the discussion. A brief summary of spectral data can be provided in the Letter as a footnote.
- Letters dealing primarily with interpretation of spectra, and those in which band shape or fine structure needs to be illustrated, may qualify for an exception
- When presentation of spectra within the paper is essential, only the pertinent sections, prepared as figures should be presented
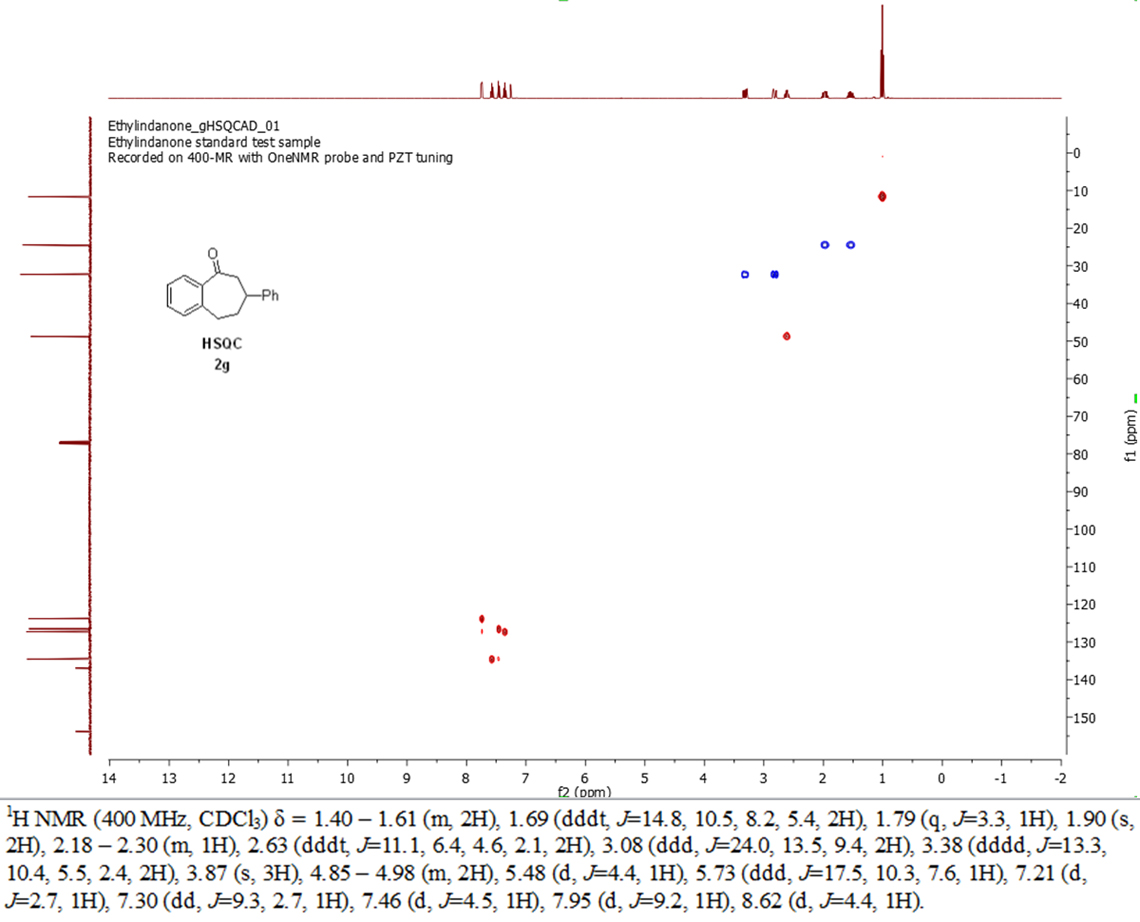
Figure 2. Typical spectroscopic data expected to support a manuscript in Organic Letters.
Full data and images of spectra should be included in the Supporting Information (see Compound Characterization and Spectra Standards for details).
So, the “User Requirements” for the submission of supplementary spectroscopic data that any author should follow are not new and give clear guidance as to the direction a solution provider must follow! Further details are given below.
The Mpublish project with Mestrelab has one major advantage in that the software partner, as with many third-party NMR analytical software providers, already has many of the tools required to deliver the formatted annotations and data rendering in their software solutions. Scientists who already work with their tools either with or without the associated databases already have installed most of the functionality required for the initial steps in the flowchart.
The additional functionality in the workflow revolves around the reporting tools and automation targeted specifically at manuscript submission.
Documentation reporting tools and packing supplementary data
These requirements are critical to getting widespread acceptance by the publishing authors as they represent an essentially “non-productive” overhead in their already stressful lives. Automation of the creation of the journal-specific peak position, coupling constant etc. information tables already saves NMR spectroscopists much effort and formatting/re-formatting in the correct manner for submission to a specific journal. Should the authors then decide to change publishers, it is even more welcome.
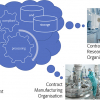
Figure 3 shows the reporting part of the solution where the specific files, loaded either from the local database or from specific individual saved files are selected before loading into the automated processing and reporting phase where the figures are generated and the entire files saved in an open document file format for later editing if required.
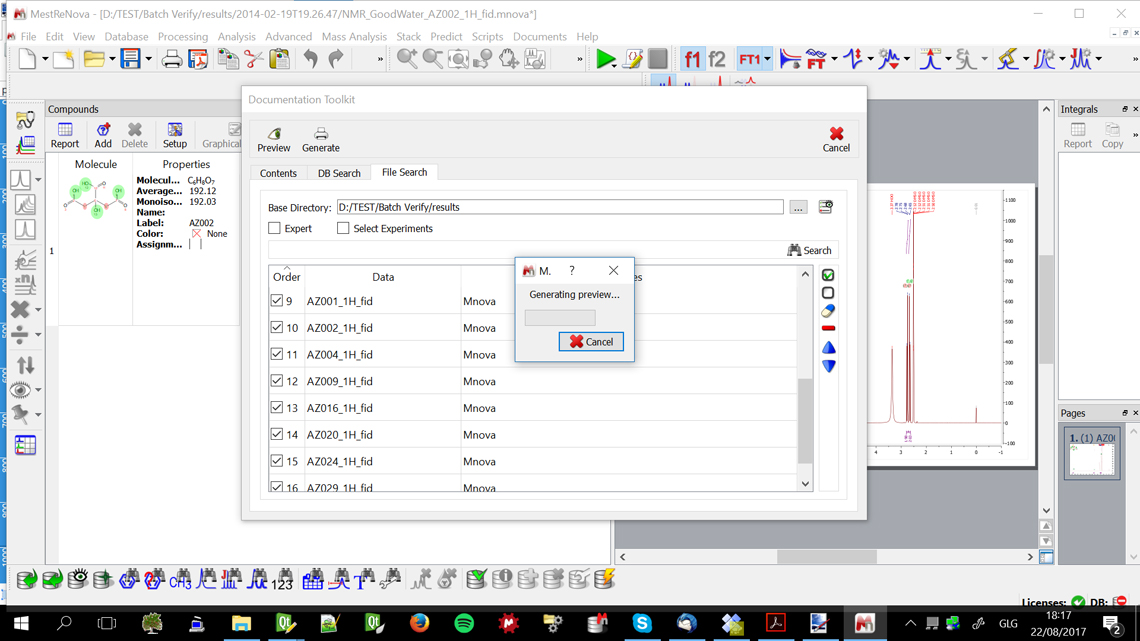
Figure 3. Automated reporting starting with the saved analytical spectra and chemical structure files.
With the figures now ready, the data packer has the capability to carry out one of the most arduous submission tasks. Organic Letters specifies in their guidance section details on how to submit the spectra as shown in the “Primary NMR data files” text box.6
Submission of primary NMR data files (FID files, acquisition data, processing parameters) is highly recommended. All original primary NMR data supporting a submission should be retained and provided if requested.
When submitting FID files:
- One folder should be created for each compound
- Folder should be named clearly, using the compound number
- Include the FID files, acquisition data and processing parameters for each experiment
- Name each spectrum according to the type of nucleus measured: 1H, 13C, DEPT, COSY, etc.
- NMR files should be compressed into zip file(s)
In a text document, include the name of the manufacturer of the spectrometer used to collect the data, the acquisition software and processing programs used to analyse the data, and the field strength used to measure each nucleus (i.e., 300 MHz 1H or 50 MHz 13C). Include a structure file that shows the structure and compound identifier for each provided dataset. MolFile is the recommended format and is strongly preferred.
I will not attempt to work out how much time this takes to do manually, but fortunately the data packer has now automated this process and, after politely asking you if you want to create a .ZIP file with all the raw data used in the document, generates the .ZIP file with individual subdirectories for each of the figures in the documents named appropriately for easy identification (Figure 4). You now have everything ready to submit to the journal with almost no additional effort over a conventional publication submission without accompanying supplementary data. Fortunately, Henry Rzepa’s data publishing workflow is much simpler but arises from a different ethos around academic institutions being considered data publishers—something to be detailed elsewhere!
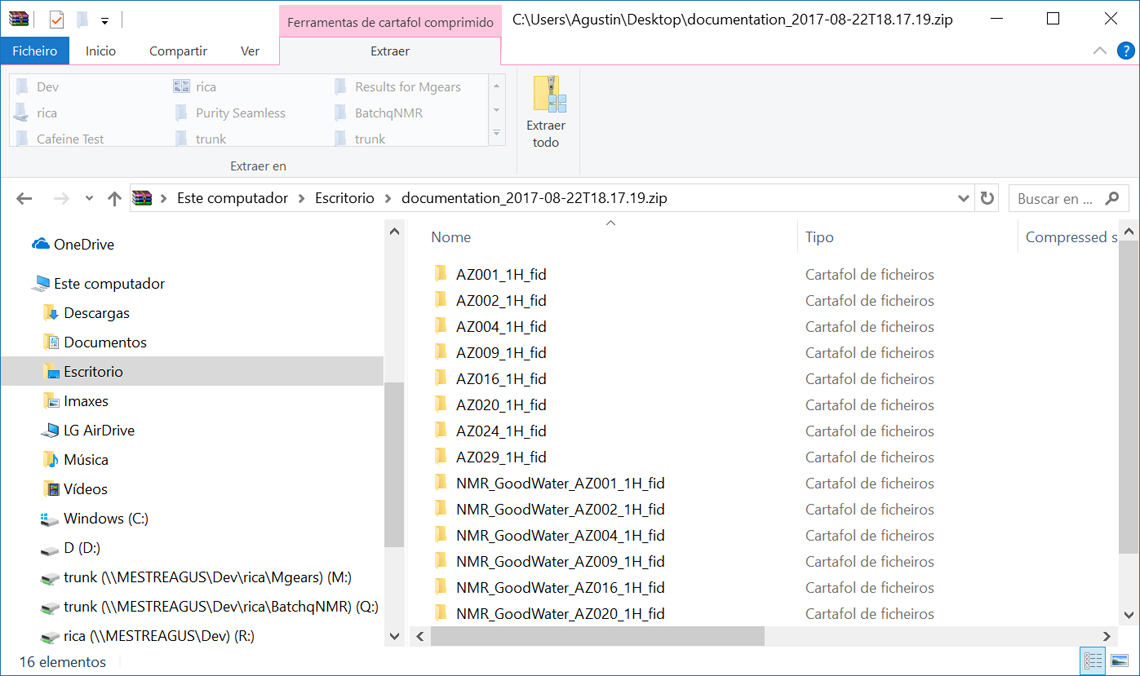
Figure 4. A zip file created as specified in the Organic Letters guidelines with separate directories for each figure in the publication.
And how to review?
So, we now only have to find a solution for the final section of the workflow. Here the generosity of the software vendors has provided a surprisingly simple solution to what could have been a nasty sticking point. As Peter Lampen rightly pointed out when reviewing this solution, it appears to be too closely linked to authors, reviewers and publishers buying a specific software product.7 As the uploaded supplementary data are in the native format of the original measuring spectrometer, it would be possible to read the files with any vendor’s solutions that are capable of parsing these raw data files. The vendor in this project has, however, come up with a nice solution.
The final stage of the workflow requires the publishers to digitally sign the submitted supplementary data file using a public/private key certification service. Reviewers are then able to download a free version of the software used to create the files in the first place and upon reading the digitally signed file, the full capability of the software is unlocked allowing the enhanced review to take place on the full supporting data. Access to the full data in this manner also strongly enhances the ability of the publishers and reviewers to spot data fraud or unwanted manipulation.
It is also worth noting that the use of the vendor software to prepare the data is not a prerequisite for the review system to work. Authors can always submit data prepared with a different processing software, upload the original raw data acquired by the instrument and could manually prepare the processed and raw data to fulfil the publisher format expectations. In this case, the raw data can still be signed by the publisher, and the free version of the review software would still allow full review of the digitally signed data.
Conclusions
It is possible for publishers and software vendors to get together to provide tools for the spectroscopic community to take most of the pain out of submitting real spectra as supplementary data. Such a solution may also be of interest within less publicly open environments, such as inside companies, as presented by Steve Hollis of Amgen and co-authors at the ENC conference in 2016 where a solution combining a number of different third-party software vendors was presented for open access NMR inside his company.8 Not all journals currently have such detailed requirements as the guidelines adopted by Organic Letters, but the solution does show that in an internet age it is possible to actually do less work whilst submitting all the full spectra supplementary data than you would need to carry out without the support of the current breed of spectroscopic data processing and reporting tools!
References
- International Union of Pure and Applied Chemistry (IUPAC), General Assembly 2017 http://www.iupac2017.0rg/assembly.php
- Access to Research Spectroscopic Data and Associated Chemical Knowledge (EUROSPEC), Project ID G7RT-CT-2001-05063. http://cordis.europa.eu/project/rcn/60180_en.html
- A.N. Davies, P. Lampen, S.R. Heller and E. Bolton, “Keeping the dream alive”, Spectrosc. Europe 27(3), 19–21 (2015). http://bit.ly/eurospec
- H.S. Rzepa, Demonstration of Professional Preview of FAIR (Findable, accessable, inter-operable and re-usable) NMR Data files using MestreNova and Mpublish (2016). https://doi.org/ 10.14469/hpc/2923. See Demonstration of Professional Preview of FAIR (Findable, accessable, inter-operable and re-usable) NMR Data files using MestreNova and Mpublish https://doi.org/10.14469/hpc/1053 for the guide to setting up and uploading files to the Imperial College Data Repository, as described by M.J. Harvey, A. McLean and H.S. Rzepa, “A metadata-driven approach to data repository design”, J. Cheminform. 9, 4 (2017). https://doi.org/10.1186/s13321-017-0190-6
- EU Guidelines to the Rules on Open Access to Scientific Publications and Open Access to Research Data in Horizon 2020, Version 3.2. 21 (March 2017). http://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-pilot-guide_en.pdf
- 2017 Guidelines for Authors, Organic Letters. http://pubs.acs.org/paragonplus/submission/orlef7/orlef7_authguide.pdf
- P. Lampen, private communication (August 2017).
- S. Hollis, D. Strand and P. Wheeler, Integrating Automation and Data Management in Open-Access NMR. ACD/Labs NMR Software Symposium at ENC 2016, Sunday, 10 April 2016.