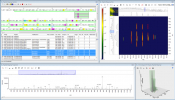
Bruker has introduced SCiLS Lab 2020 MALDI imaging software, which is now integrated with MetaboScape5.0 for automated annotations of lipids and metabolites in tissue molecular images in spatialOMx. This automatically matches ions measured on tissue to molecular information in metabolomics and lipidomics workflows, highlighting biologically relevant pathway information using MALDI imaging. The new integrated imaging and metabolomics workflow also supports data from Bruker’s scimaX™ MRMS platform, as well as from the new timsTOF fleX. MetaboScape’s T-ReX 2D algorithm performs feature extraction, de-isotoping and ion deconvolution on MALDI imaging datasets. Within MetaboScape, molecular features are annotated based on accurate mass and isotopic fidelity using SmartFormula™ and molecular information, e.g. from public databases such as HMDB and LipidMaps. MetaboScape now also offers the unique ability to increase ID scoring confidence by integrating accurate TIMS collision cross-sections (CCS) from timsTOF analyses. Identifications flow back to SCiLS Lab for fully annotated molecular images.